MA plots between VP (tipburn-damaged cultivar) vs. UH (tipburn-absent... | Download Scientific Diagram
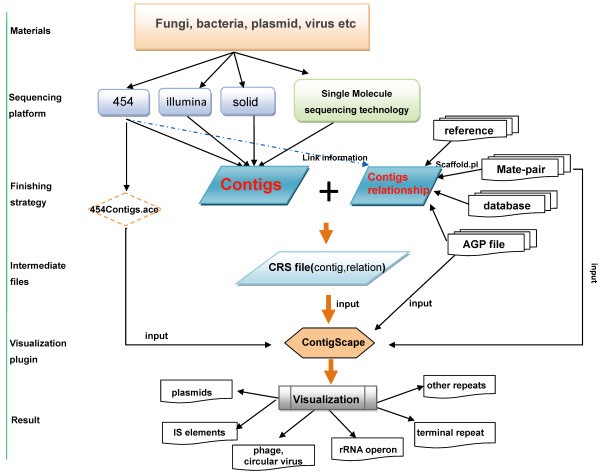
ContigScape: a Cytoscape plugin facilitating microbial genome gap closing | BMC Genomics | Full Text
Národná banka Slovenska Imricha Karvaša 1 813 25 Bratislava Vec: Oznamovacie povinnosti emitenta Vážený pán, vážená pa

Contig size distribution. x axis is the range of number of ESTs in each... | Download Scientific Diagram
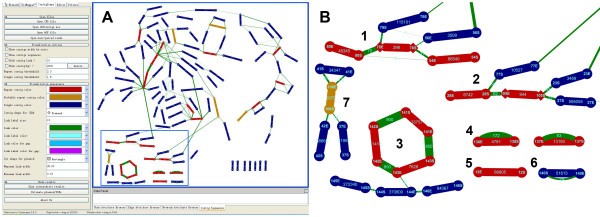
ContigScape: a Cytoscape plugin facilitating microbial genome gap closing | BMC Genomics | Full Text

Fraction of genome in haplotype blocks. X axis denote the number of... | Download Scientific Diagram
![Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity [PeerJ] Benchmarking viromics: an in silico evaluation of metagenome-enabled estimates of viral community composition and diversity [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2017/3817/1/fig-1-full.png)